Publications summarized in NIH biosketch ‘CONTRIBUTIONS TO SCIENCE’ format
1. Genomic instability in tumorigenesis and drug resistance.. Genomic instability contributes to the aggressiveness and drug resistance of lethal cancers. We are studying the plasticity of focal amplifications in mediating drug resistance, including extrachromosomal DNA (ecDNA, also known as double minutes) and homogenously staining regions (HSR). In the context of genome-wide genomic instability, our work has delineated how copy number alterations (CNA) in aneuploid cancers contribute to aggressive cancer phenotypes such as high glycolysis.
-
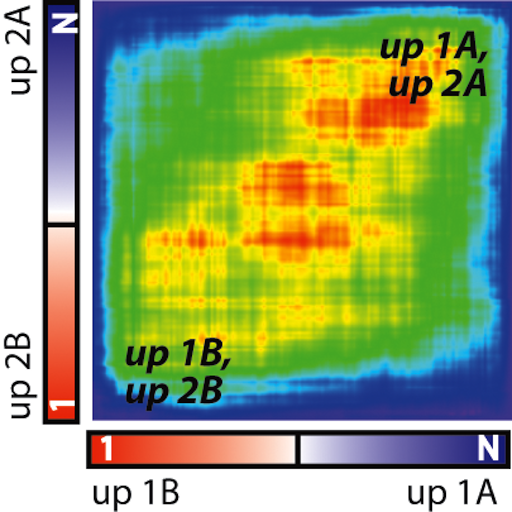
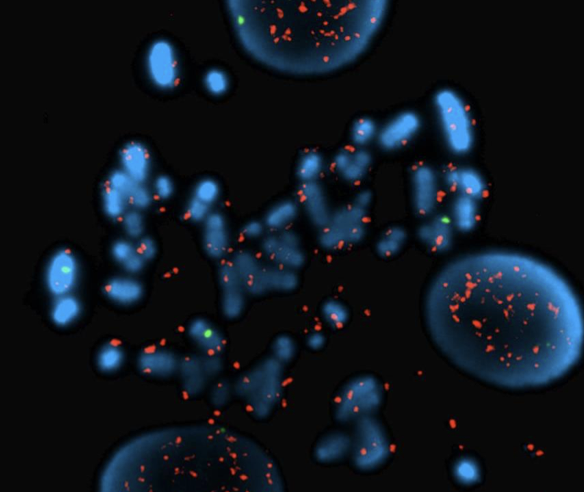 Song K, Minami JK, Huang A, Dehkordi SR, Lomeli SH, Luebeck J, Goodman MH, Moriceau G, Krijgsman O, Dharanipragada P, Ridgley T, Crosson WP, Salazar J, Pazol E, Karin G, Jayaraman R, Balanis NG, Alhani S, Sheu K, ten Hoeve J, Palermo A, Motika SE, Senaratne TN, Paraiso KH, Hergenrother PJ, Rao PN, Multani AS, Peeper DS, Bafna V, Lo RS, Graeber TG. Plasticity of extrachromosomal and intrachromosomal BRAF amplifications in mediating targeted therapy dosage challenges. Cancer Discovery, April 1, 2022: 12(4):1046-69. [article] [pdf]
Song K, Minami JK, Huang A, Dehkordi SR, Lomeli SH, Luebeck J, Goodman MH, Moriceau G, Krijgsman O, Dharanipragada P, Ridgley T, Crosson WP, Salazar J, Pazol E, Karin G, Jayaraman R, Balanis NG, Alhani S, Sheu K, ten Hoeve J, Palermo A, Motika SE, Senaratne TN, Paraiso KH, Hergenrother PJ, Rao PN, Multani AS, Peeper DS, Bafna V, Lo RS, Graeber TG. Plasticity of extrachromosomal and intrachromosomal BRAF amplifications in mediating targeted therapy dosage challenges. Cancer Discovery, April 1, 2022: 12(4):1046-69. [article] [pdf] -
Graham, N. A., Minasyan, A., Lomova, A., Cass, A., Balanis, N. G., Friedman, M., Chan, S., Zhao, S., Delgado, A., Go, J., Beck, L., Hurtz, C., Ng, C., Qiao, R., ten Hoeve, J., Palaskas, N., Wu, H., Müschen, M., Multani, A. S., Port, E., Larson, S. M., Schultz, N., Braas, D., Christofk, H. R., Mellinghoff, I. K. & Graeber, T. G. Recurrent patterns of DNA copy number alterations in tumors reflect metabolic selection pressures. Molecular Systems Biology, 2017, 13:914. [article] [pdf] [web resource: Human and mouse cancer genome copy number alteration (CNA) conservation]
-
Balanis NG, and Graeber TG. Making Mistakes Empowers Cancer Cells. Trends in Cancer 4, no. 7 [July 1, 2018]: 461-63. [pdf]
2. Prostate cancer and pan-tissue small cell neuroendocrine cancer biology. The Graeber lab is working with labs at UCLA in a pan-cancer project to characterize modes of targeted therapy resistance that involve transdifferentiation to a small cell neuroendocrine (SCN) phenotype, and can occur in prostate, lung, bladder, breast, ovarian and other cancer types. This project emerged in part from earlier work characterizing the signaling changes occurring upon progression of prostate cancer to castration-resistance.
-
Chen C-C, Tran W, Song K, Sugimoto T, Obusan MB, Wang L, Sheu KM, Cheng D, Ta L, Varuzhanyan G, Huang A, Xu R, Zeng Y, Borujerdpur A, Bayley NA, Noguchi M, Mao Z, Morrissey C, Corey E, Nelson PS, Zhao Y, Huang J, Park JW, Witte ON, Graeber TG. Temporal evolution reveals bifurcated lineages in aggressive neuroendocrine small cell prostate cancer trans-differentiation. Cancer Cell. Elsevier; 2023 Nov 22;0(0). [article] [pdf] [web resource:PARCB Time Course Multiomics Explorer]
-
 Balanis NG, Sheu KM, Esedebe FN, Patel SJ, Smith BA, Park JW, Alhani S, Gomperts BN, Huang J, Witte ON*, Graeber TG*. Pan-cancer Convergence to a Small-Cell Neuroendocrine Phenotype that Shares Susceptibilities with Hematological Malignancies. Cancer Cell. 2019 Jul 8;36(1):17-34.e7. *Corresponding authors. [article] [pdf] [web resource: Pathology Images, SCN Sample Phenotype Prediction tool, & Drug Sensitivity Prediction of SCN-like Samples tool]
Balanis NG, Sheu KM, Esedebe FN, Patel SJ, Smith BA, Park JW, Alhani S, Gomperts BN, Huang J, Witte ON*, Graeber TG*. Pan-cancer Convergence to a Small-Cell Neuroendocrine Phenotype that Shares Susceptibilities with Hematological Malignancies. Cancer Cell. 2019 Jul 8;36(1):17-34.e7. *Corresponding authors. [article] [pdf] [web resource: Pathology Images, SCN Sample Phenotype Prediction tool, & Drug Sensitivity Prediction of SCN-like Samples tool] -
BA Smith, NG Balanis, A Nanjundiah, KM Sheu, BL Tsai, Q Zhang, JW Park, M Thompson, J Huang, ON Witte*, TG Graeber*. A human adult stem cell signature marks aggressive variants across epithelial cancers. Cell Reports, 2018 Sep 18;24(12):3353-3366.e5. *Corresponding authors
-
JW Park, JK Lee, KM Sheu, L Wang, NG Balanis, K Nguyen, BA Smith, C Cheng, BL Tsai, D Cheng, J Huang, SK Kurdistani, TG Graeber*, ON Witte*. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science, 2018 Oct 5;362(6410):91–95. PMCID: in progress. *Corresponding authors. [article] [pdf] [Accompanying Perspective]
-
Drake, JM, EO Paull, NA Graham, JK Lee, BA Smith, B Titz, T Stoyanova, CM Faltermeier, V Uzunangelov, DE Carlin, DT Fleming, CK Wong, Y Newton, S Sudha, AA Vashisht, J Huang, JA Wohlschlegel, TG Graeber, Owen N Witte, & Joshua M Stuart, Patient-Specific Signaling Networks in Lethal Prostate Cancer by Phosphoproteome-Guided Multi-Omic Integration, Cell, Aug 2016, 166:1041.
-
Drake, J.M., N.A. Graham, J.K. Lee, T. Stoyanova, C.M. Faltermeier, S. Sudha, B. Titz, J. Huang, K.J. Pienta, T.G. Graeber, & Owen N. Witte. Metastatic castration-resistant prostate cancer reveals intrapatient similarity and interpatient heterogeneity of therapeutic kinase targets. Proc Natl Acad Sci U S A. 2013 Dec 3, 110(49):E4762-9. PMCID PMC3856845.
-
Drake, JM, NA Graham, T Stoyanova, A Sedghi, AS Goldstein, H Cai, DA Smith, H Zhang, E Komi-sopoulou, J Huang, TG Graeber, ON Witte. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci U S A. 2012;109:1643-1648. PMC3277127.
3. Melanoma biology and tumor immunology.The Graeber lab has applied profiling approaches to gaining leads and understanding of both melanoma cancer biology and aberrant signal transduction, as well as characterization of immune cell function in immunotherapy.
-
 Tsoi, J, L Robert, K Paraiso, C Galvan, K Sheu, J Lay, DJL Wong, M Atefi, R Shirazi, X Wang, D Braas, CS Grasso, N Palaskas, A Ribas, TG Graeber. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 2018 May 14;33(5):890-904.e5. PMCID PMC5953834. [article] [pdf] [accompanying preview] [Faculty of 1000 recommendation] [web resource: Melanoma Database Query – Create Gene Expression PCA Color Maps]
Tsoi, J, L Robert, K Paraiso, C Galvan, K Sheu, J Lay, DJL Wong, M Atefi, R Shirazi, X Wang, D Braas, CS Grasso, N Palaskas, A Ribas, TG Graeber. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 2018 May 14;33(5):890-904.e5. PMCID PMC5953834. [article] [pdf] [accompanying preview] [Faculty of 1000 recommendation] [web resource: Melanoma Database Query – Create Gene Expression PCA Color Maps] -
Titz, B, A Lomova, A Le, W Hugo, X Kong, J ten Hoeve, M Friedman, H Shi, G Moriceau, C Song, A Hong, M Atefi, R Li, E Komisopoulou, A Ribas, RS Lo, and TG Graeber. JUN dependency in distinct early and late BRAF inhibition adaptation states of melanoma. Cell Discovery, Sept 2016, 2:16028.
-
Atefi, M, B Titz, J Tsoi, E Avramis, A Le, C Ng, A Lomova, A Lassen, M Friedman, B Chmielowski, A Ribas and TG Graeber. CRAF R391W is a melanoma driver oncogene. Scientific Reports, June 2016, 6:27454. PMCID: PMC4897636.
-
Zaretsky, JM, A Garcia-Diaz, DS Shin, H Escuin-Ordinas, W Hugo, S Hu-Lieskovan, DY Torrejon, G Abril-Rodriguez, S Sandoval, L Barthly, J Saco, BH Moreno, R Mezzadra, B Chmielowski, K Ruchalski, IP Shintaku, PJ Sanchez, C Puig-Saus, G Cherry, E Seja, X Kong, J Pang, B Berent-Maoz, B Comin-Anduix, TG Graeber, PC Tumeh, TNM Schumacher, RS Lo, A Ribas. Acquired Resistance to PD-1 Blockade through Interferon Pathway Mutations. New England Journal of Medicine (NEJM), 2016 Sep, 375:819-29.
-
Hu-Lieskovan, S., S. Mok, B. Homet Moreno, J. Tsoi, L. Robert Faja, L. Goedert, E.M. Pinheiro, R.C. Koya, T.G. Graeber, B. Comin-Anduix, A. Ribas. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAFV600E melanoma. Science Translational Medicine, 2015 March 18, 7:279ra41. PMCID: PMC4765379.
-
Mueller J., O. Krijgsman, J. Tsoi, L. Robert, W. Hugo, C. Song, X. Kong, P.A. Possik, P. Cornelissen-Steijger, M. Geukes, K. Kemper, C. Goding, U. McDermott, C. Blank, J. Haanen, T.G. Graeber, A. Ribas, R.S. Lo and D.S. Peeper. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nature Communications, 2014 Dec 15, 5:5712.
-
Poulikakos, P.I., Y. Persaud, M. Janakiraman, X. Kong, C. Ng, G. Moriceau, H. Shi, M. Atefi, B. Titz, M.T. Gabay, M. Salton, K.B. Dahlman, T. Madhavi, J.A. Wargo, K.T. Flaherty, M.C. Kelley, T. Misteli, J.A. Sosman, P.B. Chapman, T.G. Graeber, A.Ribas, R.S. Lo, N.Rosen, D.B. Solit. Acquired resistance to RAF inhibitors is mediated by splicing isoforms of BRAF(V600E) that dimerize in a RAS independent manner. Nature, 2011 Nov 23, 480(7377):387-90. PMCID: PMC3266695
4. Systems biology of cancer metabolism, including mass spectrometry-based metabolomics. The Graeber lab has studied the metabolic networks disregulated in cancer, with emphasis on tying these networks to other levels of regulation such as kinase-based signaling and transcriptional regulation.
-
Xiao G, Chan LN, Klemm L, Braas D, Chen Z, Geng H, Zhang QC, Aghajanirefah A, Cosgun KN, Sadras T, Lee J, Mirzapoiazova T, Salgia R, Ernst T, Hochhaus A, Jumaa H, Jiang X, Weinstock DM, Graeber T.G., Müschen M. B-Cell-Specific Diversion of Glucose Carbon Utilization Reveals a Unique Vulnerability in B Cell Malignancies. Cell. 2018 Apr 5;173(2):470-484.e18. [article]
-
Chan, LN, Z Chen, D Braas, J-W Lee, G Xiao, H Geng, KN Cosgun, C Hurtz, S Shojaee, V Cazzaniga, H Schjerven, T Ernst, A Hochhaus, SM Kornblau, M Konopleva, MA Pufall, G Cazzaniga, GJ Liu, TA Milne, HP Koeffler, TS Ross, I Sanchez-Garcia, A Borkhardt, KR Yamamoto, RA Dickins, T.G. Graeber & M Müschen. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature. 2017 Feb 13;542(7642):479-483. [article]
-
Graham, N.A., M. Tahmasian, B. Kohli, E. Komisopoulou, M. Zhu, I. Vivanco, M.A. Teitell, Hong Wu, A. Ribas, R.S. Lo, I.K. Mellinghoff, Paul S. Mischel, and T.G. Graeber. Glucose deprivation activates a metabolic and signaling amplification loop leading to cell death. Molecular Systems Biology, 2012, 8:589. PMCID: PMC3397414. [article]
-
Palaskas, N., S.M. Larson, N. Schultz, E. Komisopoulou, J. Wong, D. Rohle, C. Campos, N. Yannuzzi, J.R. Osborne, I.Linkov, E.R. Kastenhuber, R. Taschereau, S.B. Plaisier, C. Tran, A. Heguy, H. Wu, C. Sander, M.E. Phelps, C. Brennan, E. Port, J.T. Huse, T.G. Graeber*, I.K. Mellinghoff*. 18F-fluorodeoxy-glucose positron emission tomography (18FDG-PET) marks MYC-overexpressing human basal-like breast cancers. Cancer Research, 2011, 71: 5164-74. PMC3148325. [article] (*equal contribution and co-corresponding authors).
-
Rohle, D., J. Popovici-Muller, N. Palaskas, S. Turcan C. Grommes, C. Campos, J. Tsoi, O. Clark, B. Oldrini, E. Komisopoulou, K. Kunii, A. Pedraza, S. Schalm, L. Silverman, A. Miller, F. Wang , H. Yang, Y. Chen, A. Kernytsky, M.K. Rosenblum, W. Liu, S.A. Biller, S.M. Su, C.W. Brennan , T.A. Chan, T.G. Graeber*, K.E. Yen*, I. K. Mellinghoff*. An Inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science, 2013 May 3, 340:626-30. * This work is based on equal contributions from the laboratories of T.G.G., K.E.Y., and I.K.M. PMCID PMC3985613.
-
Fu, X., R.M. Chin, L. Vergnes, H. Hwang, G. Deng, Y. Xing, M.Y. Pai, S. Li, L. Ta, F. Fazlollahi, C.Chen, R.M. Prins, M.A. Teitell, D.A. Nathanson, A. Lai, K.F. Faull, M. Jiang, S.G. Clarke, T.F. Cloughesy, T.G. Graeber, D. Braas, H.R. Christofk, M.E. Jung, K. Reue, and J. Huang. 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell Metabolism. 2015. pii: S1550-4131(15)00276-4. [Epub ahead of print]
-
Thai, M., N.A. Graham, D. Braas, M. Nehil, E. Komisopoulou, S.K. Kurdistani, Frank McCormick, TG Graeber, Heather R Christofk. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metabolism. 2014. 19(4): 694-701. PMCID PMC4294542.
5. Systems biology of cancer signaling using mass spectrometry-based proteomics. The Graeber lab helped develop the use of label-free quantitative mass spectrometry-based phosphoproteomics in studying the signaling networks driving cancer, with a particular focus on kinase inhibitor resistance mechanisms. Among other cancer topics, this work includes characterization of signaling networks in leukemia, such as that driven by Bcr-Abl, and in melanomas driven by BRAF mutations (see the related section 4 below). Data from our unbiased approaches (in this and the other sections below) has repeatedly led us to characterize feedback loops and synergies in cancer and immune biology. In our work we have pioneered using genome scale approaches (e.g. proteomics) side by side with targeted approaches (e.g. antibody-based) in iterative experimentation to simultaneously build a network-scale and a molecularly detailed view of biological processes.
-
Rubbi, L., B. Titz, L. Brown, E. Galvan, E. Komisopoulou, S.S. Chen, T. Low, M. Tahmasian, B. Skaggs, M. Müschen, M. Pellegrini, T. G. Graeber. Global phosphoproteomics reveals crosstalk between Bcr-Abl and negative feedback mechanisms controlling Src signaling. Science Signaling, 2011, 4(166):ra18. PMCID PMC4057100.
-
Titz, B., T. Low, E. Komisopoulou, S. Chen, L. Rubbi, T.G. Graeber. The proximal signaling network of the BCR-ABL1 oncogene shows a modular organization. Oncogene, 2010, 29: 5895-5910.
- Skaggs, B., M. Gorre, A. Ryvkin, M. Burgess, Y. Xie, Y. Han, E. Komisopoulou, L.M. Brown, J.A. Loo, E.M. Landaw, C.L. Sawyers, and T.G. Graeber. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc. Natl. Acad. Sci. USA, 2006 December 19, 103(51): 19466.
6. Bioinformatic algorithms – development and application. The Graeber lab has developed novel algorithms and approaches for characterizing molecular profiling data, and for integrating such data with additional data sources such as data bases and the literature. Many of these approaches were first developed in cancer projects, but have found applications in other areas such as immune function and brain development. These bioinformatic approaches continue to be used in our lab, and in publications from other groups. We provide web-based implementations of our algorithms. Three algorithm/theoretical manuscripts are listed below (a-c). Furthermore, we have a long track record of successful implementation of bioinformatic and computational approaches in cancer and immune biology discovery and classification. These implementations are part of the vast majority of our independent and collaborative manuscripts, including two of the manuscripts below (d-e).
-
Plaisier S.B., R. Taschereau, J.A. Wong, and T.G. Graeber. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Research, September 2010, 38(17): e169. PMCID: PMC2943622. Web-based implementation accessible at http://systems.crump.ucla.edu/rankrank/.
-
Graeber, T.G., and D. Eisenberg. Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nature Genetics, 2001 November, 29(3):295-300.
-
Graeber, T.G., J.R. Heath, B.J. Skaggs, M.E. Phelps, F. Remacle and R. D. Levine, Maximal Entropy Inference of Oncogenicity from Phosphorylation Signaling. Proc Natl Acad Sci U S A, March 2010, 107(13), 6112-6117. PMCID: PMC2851899.
-
D.J. Mulholland, L.M. Tran, Y. Li, H. Cai, A. Morim, S. Wang, S. Plaisier, I.P. Garraway, J. Huang, T.G. Graeber, Hong Wu. Cell Autonomous Role of PTEN in Regulating Castration-Resistant Prostate Cancer Growth. Cancer Cell, 2011, 19(6):792-804. (Bioinformatic contribution)
- Ellwood-Yen, K., T.G. Graeber†, J. Wongvipat, M. Luisa-Aripse, J. Zhang., R. Matusik, G.V. Thomas and C.L. Sawyers. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell, 2003 September 4, 4(3):223-38. (†computational biology contribution)
7. Skin disease and immunology bioinformatics & integrative genomics. Through the development of bioinformatic approaches the Graeber lab has worked to use transcriptome and miRNA profiling data to gain insight into the dysregulation present in diseased and infected skin tissue, and into the functional aspects of the spectrum of immune cells that combat the infections.
-
Teles, R.M.B., T.G. Graeber*, S.R. Krutzik, D.Montoya, M.Schenk, D.J. Lee, E. Kelly-Scumpia, R. Chun, E.N. Sarno, T.H. Rea, M. Hewison, J.S. Adams, B.R. Bloom, G. Cheng, R.L. Modlin. Type I IFN suppresses Type II IFN triggered human antimicrobial responses. Science, 2013, 339:1448-53. PMCID: PMC3653587. (Bioinformatic and data-analysis contribution)
-
Liu, P.T., M. Wheelwright, R. Teles, E. Komisopoulou, K. Edfeldt, B. Ferguson, A. Vazirnia, T. Rea, E.N. Sarno, T.G. Graeber, and R.L. Modlin. Induction of miRNAs during leprosy infection inhibits a host antimicrobial pathway. Nature Medicine, 2012;18:267-73. PMC3274599. (Bioinformatic contribution).
-
Schenk M., S.R. Krutzik, P.A. Sieling, D.J. Lee, R.M.B. Teles, M.T. Ochoa, E.N. Sarno, T.H. Rea, T.G. Graeber, S. Kim, G. Cheng and R.L. Modlin. NOD2 triggers an IL-32 dependent human dendritic cell program in leprosy. Nature Medicine, 2012 Mar 25;18(4):555-63. PMC3348859 (Bioinformatic contribution).
-
Bleharski J.R.*, H. Li*, C. Meinken*, T.G. Graeber*, M.-T. Ochoa, M. Yamamura, A. Burdick, E.N. Sarno, M. Wagner, M. Röllinghoff, T.H. Rea, M. Colonna, S. Stenger, B.R. Bloom, D. Eisenberg and R.L. Modlin. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science, 2003 September 12, 301(5639):1527-30. (*equal contribution)
8. Identification of hypoxia as a physiological selective force for p53 mutation. The findings from the ressearch I led as a graduate student provided an explanation for why p53 is mutated in a high percentage of primary tumors. This work is covered by many reviews (Hanahan & Weinberg, The hallmarks of cancer, Cell 100:57) and textbooks (Robbins pathologic basis of disease, 1999).
-
Olcina MM, Balanis NG, Kim RK, Aksoy BA, Kodysh J, Thompson MJ, Hammerbacher J, Graeber TG, Giaccia AJ. Mutations in an Innate Immunity Pathway Are Associated with Poor Overall Survival Outcomes and Hypoxic Signaling in Cancer. Cell Reports. 2018 Dec 26;25(13):3721-3732.e6. PMID: 30590044. [article]
-
Graeber, T.G., C. Osmanian, T. Jacks, D.E. Housman, C.J. Koch, S.W. Lowe and A.J. Giaccia. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature, 1996 January 4, 379(6560):88-91. (cited over 2350 times; accompanying News and Views) [article] [news & views]
-
Graeber, T.G., J.F. Peterson, M. Tsai, K. Monica, A.J. Fornace Jr. and A.J. Giaccia. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Molecular and Cellular Biology, 1994, 14(9):6264-6277. PMCID: PMC359153
Complete list of published work online in NCBI MyBibliography:
https://www.ncbi.nlm.nih.gov/myncbi/thomas.graeber.1/bibliography/public